Gene KO by genome-wide KO double genetic essentiality screen in NALM6 cells
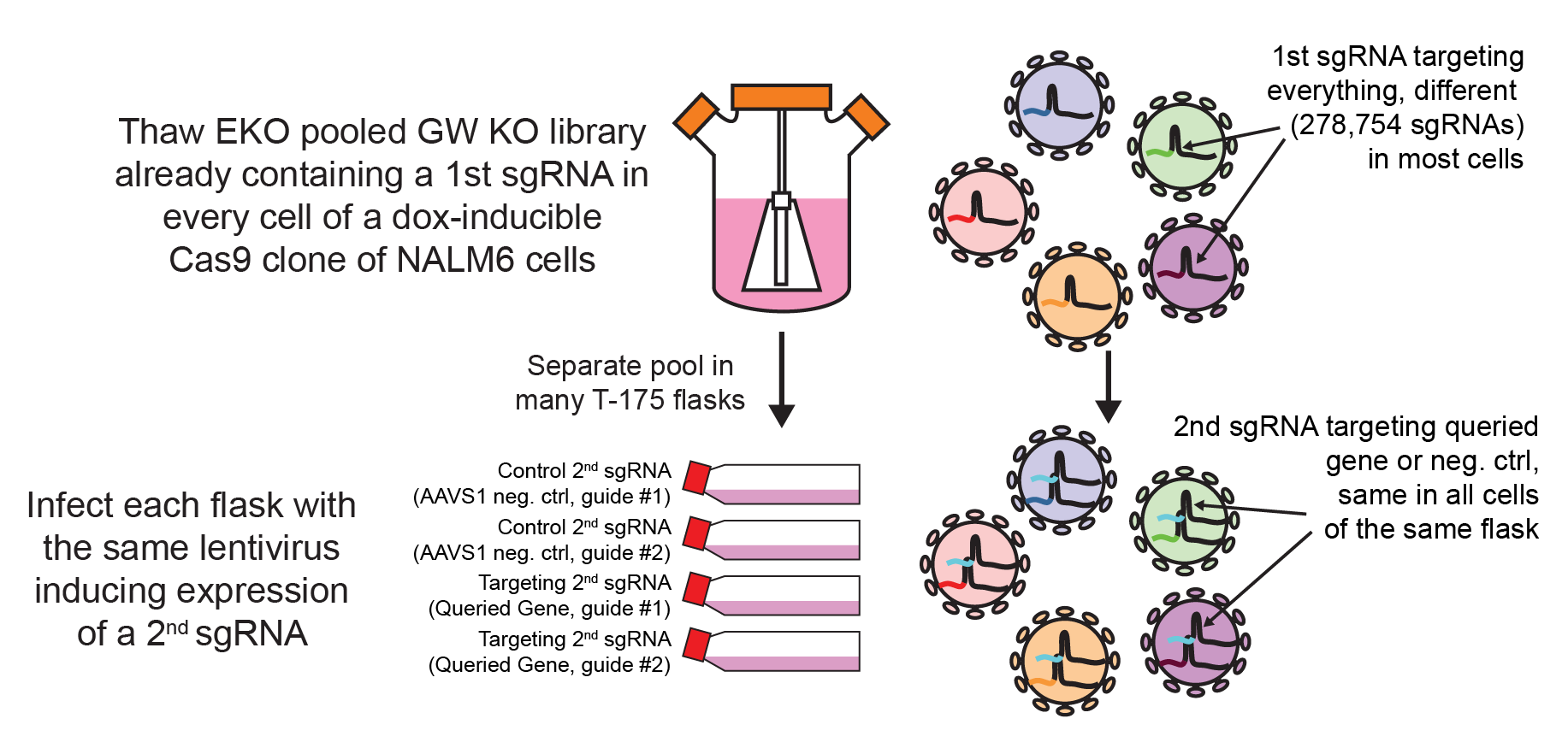
We offer genome-wide double knock-out screens. For each query gene you select we will take care to
clone 2 independent sgRNAs sequences targeting that gene and make lentivirus from them. We will
use them independently to infect and select our genome-wide (GW) KO pooled library in NALM-6
cells (2 sgRNA per cell: 1 sgRNA per cell from the GW pool and 1 sgRNA against the queried gene).
After Cas9 induction for 7 days and outgrowth for 8 days, we recover genomic DNA, PCR-amplify
sgRNA sequences from the GW pool, and score gene essentiality comparing the sample with the controls
(cells infected with non-targeting sgRNAs).
Academia: $5,200 CAD per gene
Industry: please contact for a quote
Double genetic screening
Double KO experiments allow the identification of genes crucial for cellular fitness in homeostatic or
stress conditions. In fact, single gene knockouts that do not show any phenotype can display a significant
decrease or increase of cellular fitness when deleted together (called synthetic lethality). While this
approach has been extensively used in yeast to define a genome-scale genetic interaction map
(PMID:20093466),
because of technical hurdles, it has never been performed on a genome-wide scale in human.
ChemoGenix has designed a simple yet powerful approach that allows to test a single gene vs the whole genome.
We infect our GW-KO library with a lentivirus encoding an sgRNA targeting the query gene. As a result, every
cell in the pool, prior to Cas9 induction, will contain 2 sgRNAs, one from the library and one targeting the
query gene. This method has 2 main advantages, it doesn’t require the generation of a costly clonal KO line
and, because the induction of Cas9 happens only once both guides are inside the cells, it allows testing
of essential genes. We routinely test 2 guides per gene to mitigate noise.
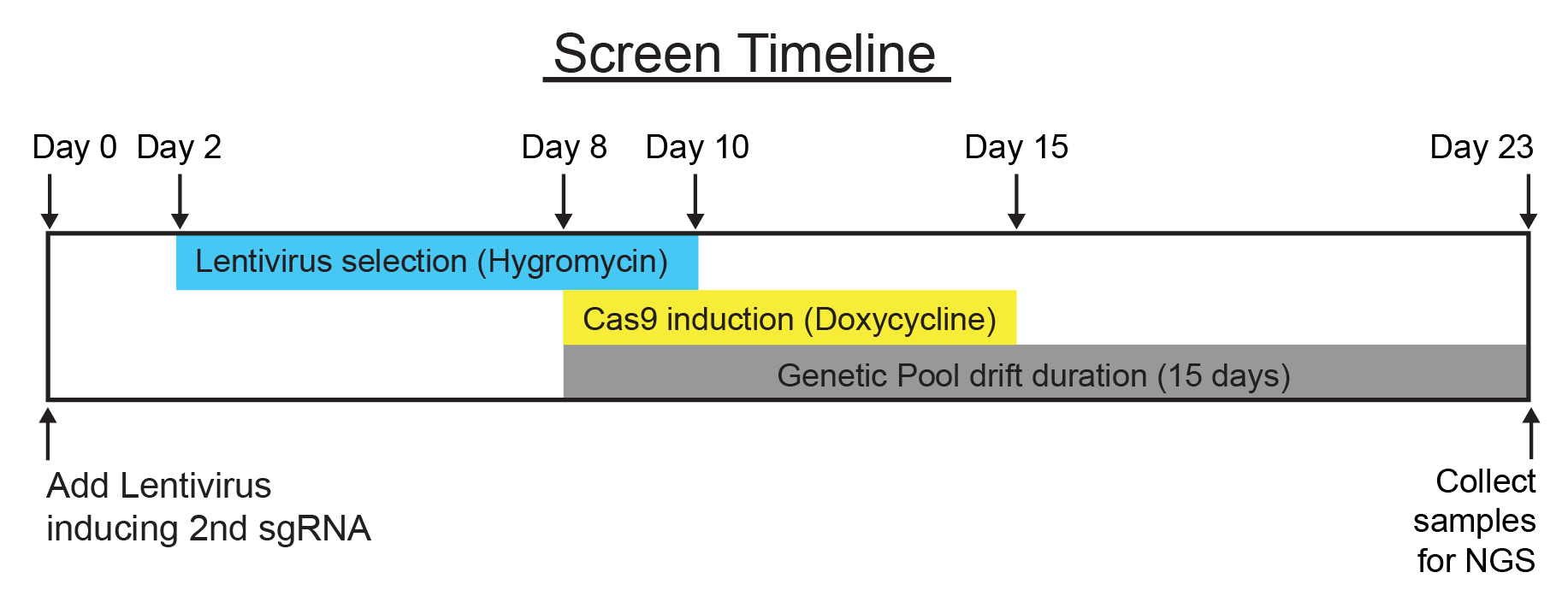
Because we need to infect and select our GW library with the lentivirus encoding the 2nd sgRNAs prior to Cas9 induction with doxycycline, this adds 8 more days of culture. Following hygromycin selection, cells are cultured for 15 days of population drift (7 days dox, 8 days without).
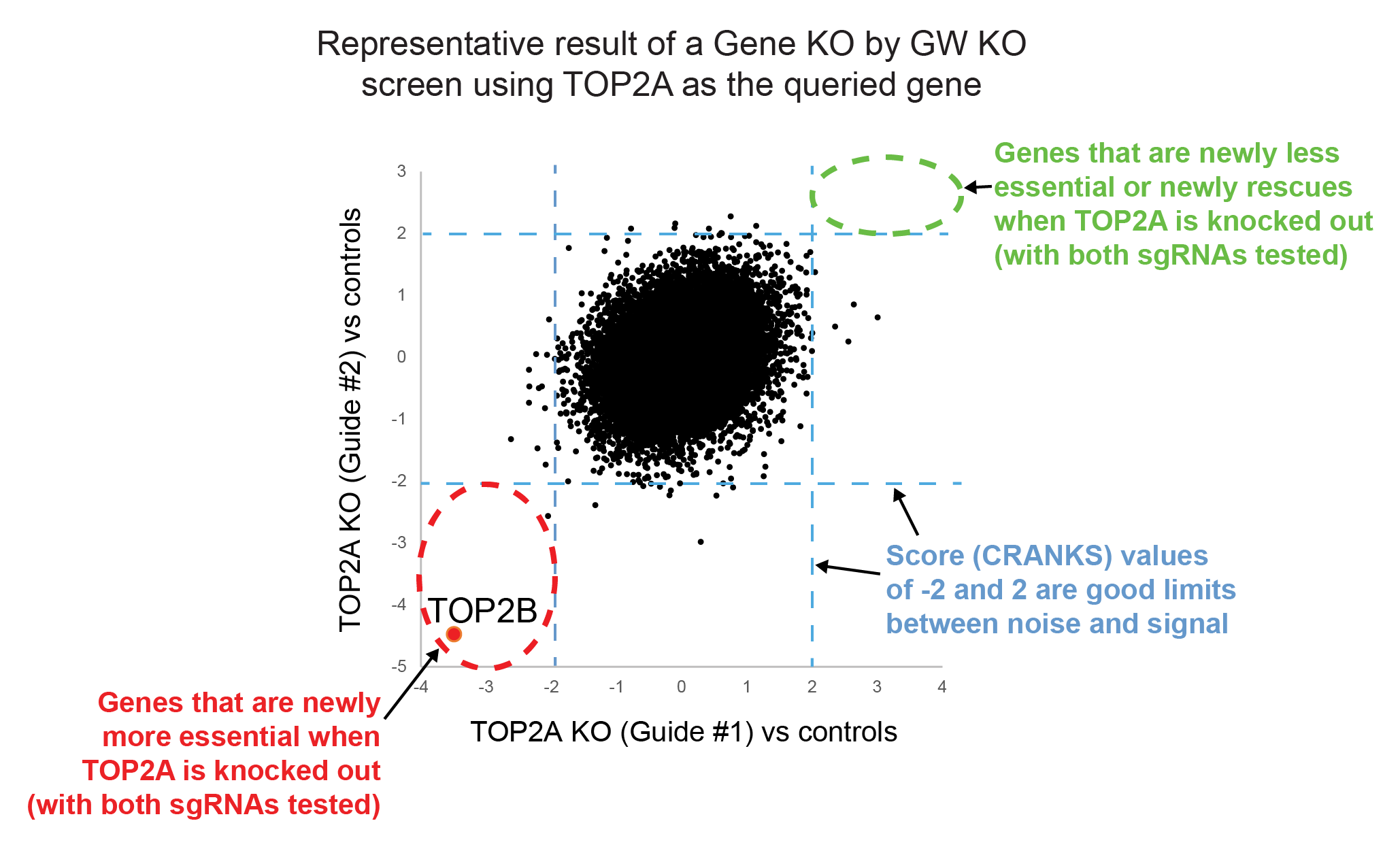
Below, you can see a typical result from a Gene KO by GW KO screen made in NALM6 cells using TOP2A KO as a query gene. Topoisomerase 2 disentangles chromosomes and is a heterodimer formed from TOP2A and TOP2B gene products. The results show that when we knock out TOP2A, TOP2B is the gene whose essentiality has increased the most. There are a few publications that indicate that Topo II also exists as a homodimer of either TOP2A or TOP2B and t.his data shows the essential role of TOP2B homodimer when TOP2A is deleted.
Data Confidentiality
The findings from academic and non-profit clients will be made publicly accessible on this website three years
after their completion. However, this provision does not extend to for-profit clients, who retain exclusive
ownership of their data.
Contact us
Please contact us in order to discuss your needs and to take advantage of the most appropriate solution.