CRISPR Screen Basics
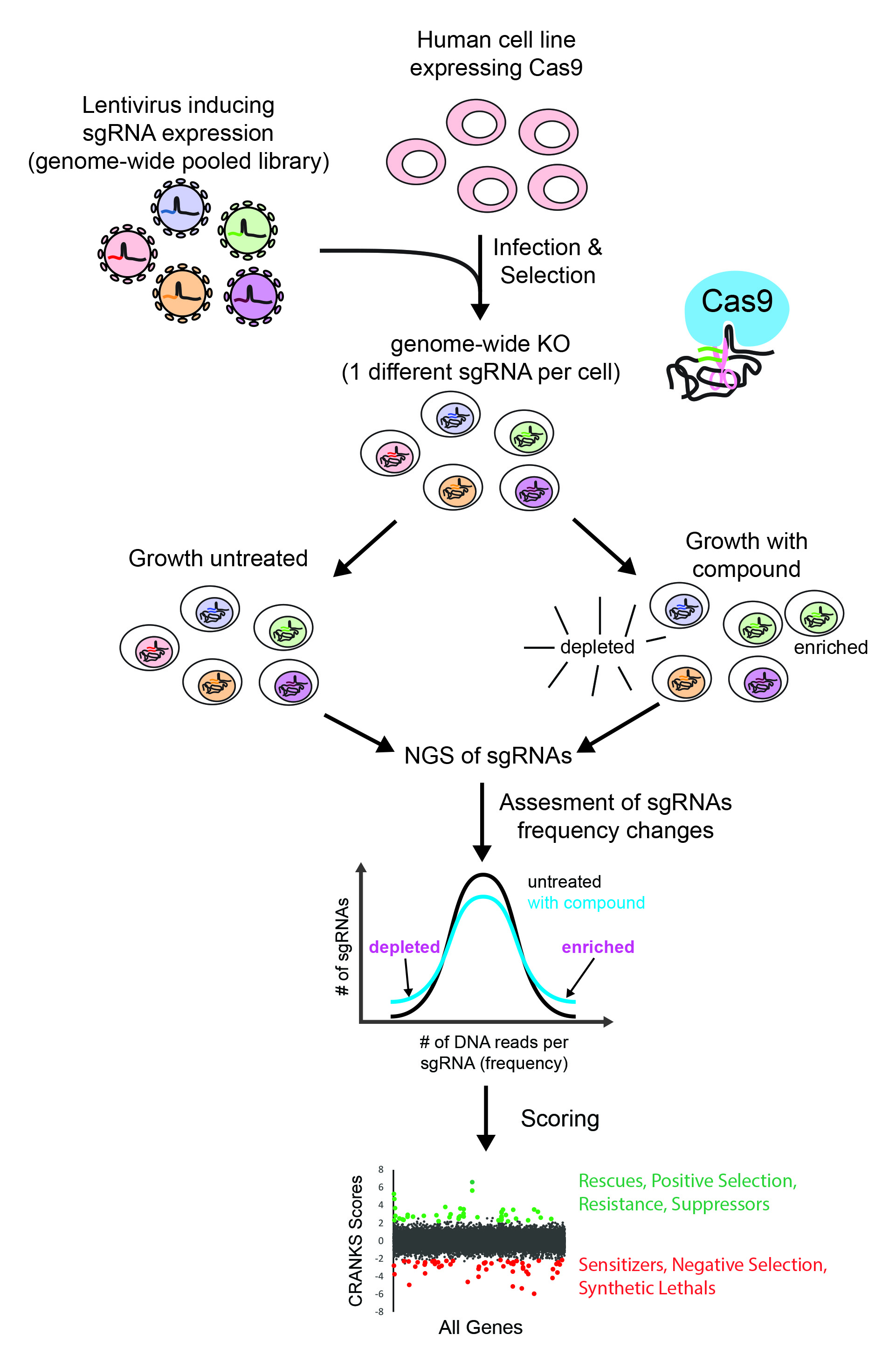
The advent of CRISPR/Cas9 technology has significantly transformed gene-editing capabilities, enabling comprehensive functional genome-wide screenings directly in human cells. The Cas9 endonuclease from Streptococcus pyogenes can be precisely guided to induce double-strand breaks (DSBs) at specific DNA sites through a 20-base pair interaction with a single-guide RNA (sgRNA). Following the initial cut, the DNA undergoes repair processes, which may lead to further cuts until non-homologous end-joining repairs result in insertions or deletions (indels). This often results in a frame-shift mutation within the targeted gene's exon, effectively inactivating the gene product and producing a functional knockout.
Utilizing pooled knockout sgRNA libraries allows researchers to interrogate the entire genome in a single experiment. These libraries consist of hundreds of thousands of sgRNA molecules, each cloned into lentiviral plasmids, with one sgRNA per plasmid, all contained within a single tube. By infecting cells at a low multiplicity of infection (MOI), the delivery of a single sgRNA per cell is encouraged. When these cells are cultured in the presence of a growth-inhibiting chemical, those harboring sgRNAs that confer resistance or sensitivity to the drug will be either enriched or depleted in the population, respectively.
The variations in sgRNA frequencies, analyzed through Next-Generation Sequencing (NGs), allow for the calculation of robust enrichment or depletion scores for all genes. The identified genes, categorized as rescues (knockouts that confer resistance, leading to sgRNA enrichment and positive scores) or synthetic lethals (knockouts that induce sensitivity, resulting in sgRNA depletion and negative scores), collectively represent the chemogenomic signature associated with a specific compound.